Choose Cell Lines for Endometrial Cancer Research: Harvard Sander Lab
RL95-2, HEC-1, AN3-CA, MFE-296, and SNG-M are some of the most widely used cell lines for endometrial cancer research. Often researchers have questions on selecting the most appropriate cell line(s) for particular research uses.
TumorComparer utilizes several data types (e.g., mutation, copy number, and expression) to perform a comparison of The Cancer Genome Atlas (TCGA) tumor data to experimental cell line models from the Cancer COSMIC Cell Line Project (CCLP) for several cancer types. Please refer to Sinha R et al., 2021. DOI: 10.1016/j.crmeth.2021.100039 for details on the methodology.
An analysis for endometrial cancer cell lines with differences between the endometrial cancer cell lines RL95-2 vs HEC-1; RL95-2 vs AN3-CA; RL95-2 vs MFE-296; and etc.
If you use content from this page, please cite Sinha R et al., 2021. DOI: 10.1016/j.crmeth.2021.100039.
Characteristics of Endometrial Cancer Cell Lines
The figure shows the presence of alterations for genes (ARID1A, FBXW7, FGFR2, PTEN, IGF1R, etc.) commonly associated with endometrial cancer in the cell lines: EN, MFE319, AN3CA, MFE296, SNGM, etc. The gene list is retrieved from DisGeNET, and the alteration information is retrieved from cBioPortal for the Cancer Cell Line Encyclopedia (CCLE) dataset.
NOTE: Only a subset of endometrial cancer cell lines are shown to ensure a readable figure. Alterations for additional endometrial cancer cell lines and genes are available on cBioPortal using links on the table below.
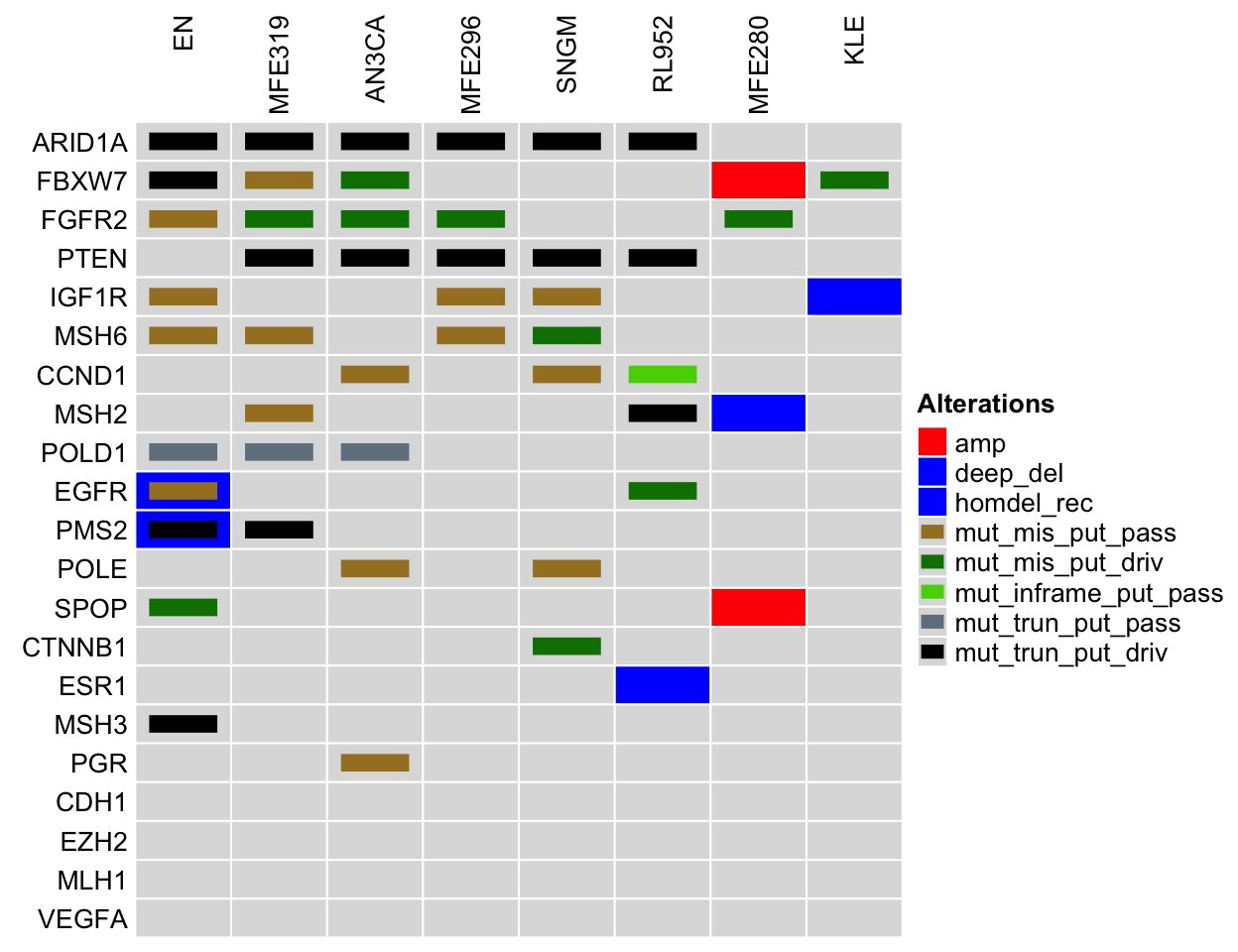
Comparing endometrial cancer cell lines to tumor samples. Legend Abbreviations: mrna_hi = mRNA High; mrna_lo = mRNA Low; prot_hi = Protein High; prot_lo = Protein Low; mut_mis_put_pass = Missense Mutation (putative passenger); mut_mis_put_driv = Missense Mutation (putative driver); mut_inframe_put_pass = Inframe Mutation (putative passenger); mut_inframe_put_driv = Inframe Mutation (putative driver); mut_trun_put_pass = Truncating mutation (putative passenger); mut_trun_put_driv = Truncating mutation (putative driver); amp = Amplification; deep_del = Deep Deletion; homdel_rec = homdel_rec; amp_rec = amp_rec; sv = sv; sv_rec = sv_rec; splice = splice
List of Endometrial Cancer Cell Lines Compared to Pan-Cancer TCGA
The TumorComparer analysis examines the most variably expressed genes, as well as alteration information for cell lines and TCGA patient data using a weighted similarity approach; for details, read: Sinha R et al., 2021. DOI: 10.1016/j.crmeth.2021.100039.
More information for the endometrial cancer cell lines is available on cBioPortal using links on the table below.
| Cell Line | % Rank by Avg % Ranks | More Line Info | % Rank by Mutation | % Rank by Copy Number | % Rank by Expression |
|---|---|---|---|---|---|
| MFE319 | 0.92 | Link | 0.35 | 0.97 | 1.00 |
| COLO684 | 0.90 | Link | 0.36 | 0.99 | 0.89 |
| RL952 | 0.89 | Link | 0.50 | 0.97 | 0.73 |
| MFE280 | 0.88 | Link | 0.64 | 0.53 | 0.99 |
| AN3CA | 0.86 | Link | 0.64 | 0.60 | 0.89 |
| KLE | 0.85 | Link | 0.86 | 0.37 | 0.89 |
| EN | 0.50 | Link | 0.52 | 0.05 | 0.92 |
| SNGM | 0.50 | Link | 0.59 | 0.01 | 0.92 |
| HEC1 | 0.42 | NA | 0.34 | 0.25 | 0.80 |
| MFE296 | 0.38 | Link | 0.62 | 0.01 | 0.69 |
Cell Lines with Common Mutations Found in TCGA Endometrial Cancer
Alterations present in cell lines also present in at least two TCGA Pan-Cancer Atlas (UCEC) samples on cBioPortal.
Alterations
PTEN F341V (TCGA Altered: 1%; 5 out of 509 samples)
- JHUEM7
PTEN R130Q (TCGA Altered: 6.7%; 34 out of 509 samples)
- AN3CA
- JHUEM1
- MFE296
PTEN R142W (TCGA Altered: 2%; 10 out of 509 samples)
- ESS1
PTEN R173H (TCGA Altered: 0.8%; 4 out of 509 samples)
- RL952
FGFR2 C382R (TCGA Altered: 1%; 5 out of 509 samples)
- JHUEM2
FGFR2 N549K (TCGA Altered: 1.4%; 7 out of 509 samples)
- AN3CA
- MFE296
FGFR2 S252W (TCGA Altered: 4.7%; 24 out of 509 samples)
- MFE280
- MFE319
SPOP R121Q (TCGA Altered: 0.6%; 3 out of 509 samples)
- EN
SPOP S14L (TCGA Altered: 0.6%; 3 out of 509 samples)
- HEC108
FBXW7 R465C (TCGA Altered: 1.6%; 8 out of 509 samples)
- ESS1
FBXW7 R465H (TCGA Altered: 2%; 10 out of 509 samples)
- JHUEM3
FBXW7 R479Q (TCGA Altered: 1%; 5 out of 509 samples)
- HEC251
- KLE
FBXW7 R505C (TCGA Altered: 1.6%; 8 out of 509 samples)
- JHUEM1
POLE A456P (TCGA Altered: 0.4%; 2 out of 509 samples)
- ESS1
POLE P286R (TCGA Altered: 3.9%; 20 out of 509 samples)
- HEC251
- JHUEM7
CTNNB1 S37C (TCGA Altered: 2%; 10 out of 509 samples)
- JHUEM2
CTNNB1 S37P (TCGA Altered: 0.4%; 2 out of 509 samples)
- HEC108
- SNGM
Customized Comparisons
Each project has unique questions. TumorComparer (tumorcomparer.org) facilities customized comparisons of cell line datasets to tumor samples directly from the browser.
Technical Information
This analysis was generated using TumorComparer: 0.99.25. If you use content from this page, please cite Sinha R et al., 2021. DOI: 10.1016/j.crmeth.2021.100039.